GRPhIN: Graphlet Characterization of Regulatory and Physical Interaction Networks
computational biology, bioinformatics, data science, network analysis, algorithms, AI automation, health informatics, biotechnology research
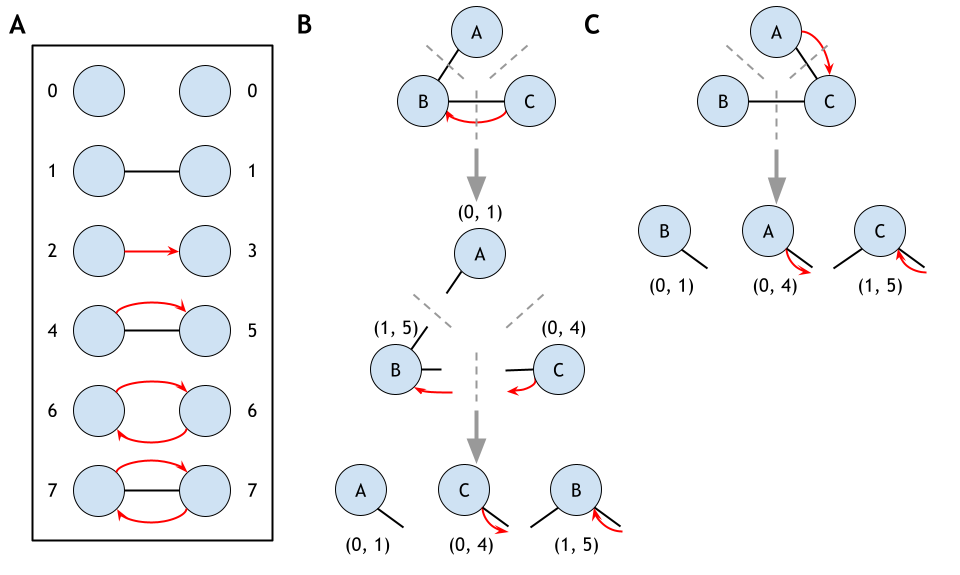
GRPhIN (Graphlet Characterization of Regulatory and Physical Interaction Networks) is an algorithm for counting graphlets and the specific node positions within each graphlet (called orbits) in mixed regulatory and physical interaction networks. Graph representions of regulatory or physical interactions in isolation may obscure the complete functional context of a protein. PPI networks and GRNs do not exist separately; proteins are transcription factors, genes encode proteins, and physical and regulatory interactions mix and coexist forming their own distinct patterns. Graphlets are small, connected, induced subnetworks that describe patterns, local topologies, and organization in networks.
GRPhIN takes as input (1) an undirected PPI network and (2) a directed regulatory network and counts all mixed graphlets and their respective orbits (Figure 6). GRPhIN provides additional functional context to the roles a protein may play beyond traditional isolated network types.
Learn more about GRPhIN at the GitHub Repository or read the preprint here. GRPhIN was selected as a full length talk at GLBIO 2025.
I am grateful for the opportunity to have worked with Altaf Barelvi and Dr. Anna Ritz on this project and get hands on experience developing algorithms.
Oliver F. Anderson, MS – Computational Biologist, Data Scientist, and Research Consultant based in Portland, Oregon. I design data-driven solutions in bioinformatics, machine learning, and AI automation for research and biotech.
Back to top