GRPhIN: Graphlet Characterization of Regulatory and Physical Interaction Networks
GRPhIN, graphlets, regulatory networks, protein-protein interaction, network motifs, mixed networks, orbits, algorithms, bioinformatics, graph theory
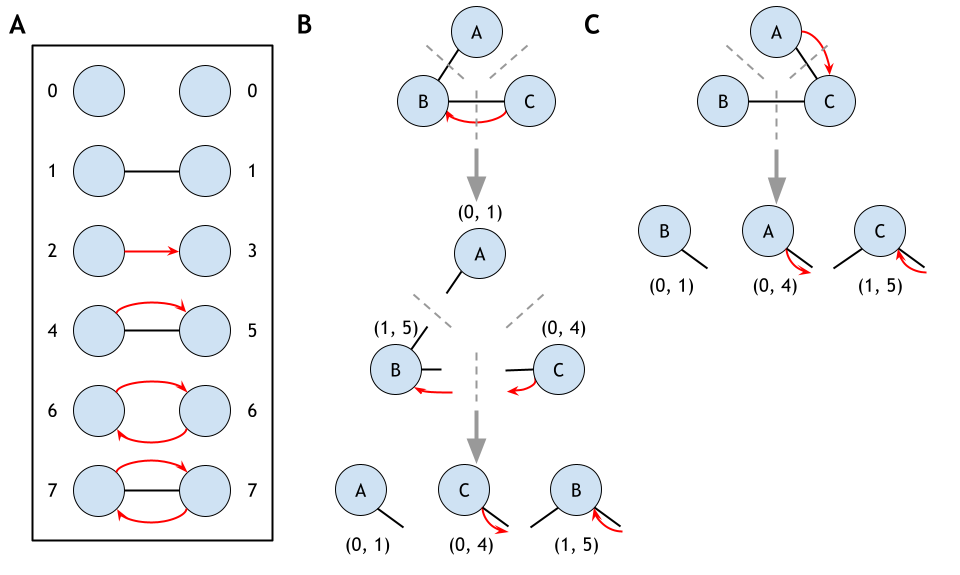
GRPhIN (Graphlet Characterization of Regulatory and Physical Interaction Networks) is an algorithm designed to count small subnetworks, known as graphlets, and the specific node positions within them, called orbits. Unlike isolated protein–protein interaction (PPI) networks or gene regulatory networks (GRNs), GRPhIN captures how regulatory and physical interactions coexist and form distinct structural patterns. This provides a richer functional context for understanding the roles of proteins in complex systems.
You can access the code on GitHub or read the paper in Bioinformatics Advances here. GRPhIN was selected as a full-length talk at GLBIO 2025.
Workflow
- Input networks
- Undirected protein–protein interaction (PPI) network.
- Directed regulatory network.
- Graphlet enumeration
- Identify all 2- and 3-node mixed graphlets (regulatory, physical, and both edges).
- Assign unique identifiers to each graphlet type.
- Orbit labeling
- Label node positions (orbits) within each graphlet.
- Capture the distinct roles proteins may play depending on interaction directionality.
- Sorting & uniqueness
- Apply orbit sorting to ensure consistent labeling.
- Generate canonical representations of graphlets across isomorphic structures.
- Output
- Counts of all graphlets and their associated orbits in the mixed network.
- Data exportable for downstream statistical or comparative analysis.
Discussion
By integrating regulatory and physical interactions, GRPhIN reveals structural patterns that are invisible when networks are analyzed separately. The algorithm highlights how proteins can act in multiple contexts. For example, proteins may serve as both transcriptional regulators and participants in physical complexes.
GRPhIN provides: - Functional insight into proteins beyond a single network type. - The ability to compare structural motifs across organisms or experimental conditions. - A foundation for testing hypotheses about protein function in mixed interaction settings.
The method is already being used to benchmark motif counts across organisms and has applications in studying pathway regulation, protein complexes, and evolutionary comparisons.
This work resulted in a publication and a full-length talk:
- Presented at the GLBIO 2025 conference
- Published in: 👉Bioinformatics Advances
FAQ: Using GRPhIN
What is a graphlet?
Graphlets are small, connected subnetworks (2–5 nodes) that represent recurring topological patterns. GRPhIN currently focuses on 2- and 3-node graphlets in mixed interaction networks.
Why combine regulatory and physical interactions?
Analyzing PPI or regulatory networks alone misses the ways proteins act across interaction types. GRPhIN integrates both to capture the full range of structural roles proteins play.
What are orbits?
Orbits define the unique positions a node can occupy within a graphlet. By tracking orbits, GRPhIN characterizes the specific roles of proteins across different graphlet structures.
Where can I access GRPhIN?
GRPhIN is open source on GitHub. A detailed description is available in the publication.
I am grateful for the opportunity to have worked with Altaf Barelvi and Dr. Anna Ritz on this project and get hands-on experience developing algorithms.
Oliver F. Anderson, MS – Computational Biologist, Data Scientist, and Research Consultant based in Portland, Oregon. I design data-driven solutions in bioinformatics, machine learning, and AI automation for research and biotech.
Back to top